Highlights
- Identification of five key pathogenic variants (ATM, BRCA2, CHEK2, HOXB13, PALB2) significantly associated with aggressive prostate cancer risk in men of African ancestry.
- Integration of rare germline variants, polygenic risk scores (PRS), and family history refines lifetime absolute risk estimates for prostate cancer, including aggressive and metastatic forms.
- Substantial risk variability supports moving beyond race-based screening guidelines toward personalized screening approaches based on genetic risk stratification.
- Early detection strategies focusing on high-risk individuals could improve outcomes and reduce overdiagnosis in African ancestry populations.
Introduction
Prostate cancer (PCa) exhibits high heritability, with genetic factors accounting for approximately 58% of risk variability. While common genetic variants underpin risk prediction through polygenic risk scores (PRS), rare germline pathogenic variants (PVs) in DNA repair and cancer predisposition genes contribute significantly to aggressive and early-onset disease. Most existing studies have focused on European ancestry populations, leaving a critical knowledge gap regarding genetic risk factors and their clinical implications in men of African ancestry. Given the disproportionately high PCa incidence and mortality within African ancestry groups, there is an urgent need to characterize the genetic risk spectrum to inform personalized screening and management strategies that transcend the limitations of race-based guidelines.
Study Design and Methods
This large multi-national case-control study involved 7,176 prostate cancer cases and 4,873 controls of African ancestry recruited from seven countries across North America and Africa. Cases included nonaggressive, intermediate-risk, aggressive, and metastatic PCa subtypes, defined by pathological stage, Gleason score, PSA levels, and mortality data. Whole-exome sequencing was performed to identify rare PVs across 37 cancer predisposition genes, with a focus on 16 panel genes commonly used in clinical testing. PRS were calculated using 451 validated PCa risk variants derived from multi-ancestry genome-wide association studies, and family history information was incorporated.
Gene-based associations were assessed using logistic regression adjusted for age and genetic ancestry. PVs were classified using standard variant effect predictors and ClinVar annotations. Lifetime absolute risk estimates, accounting for PV status, PRS, and family history, were generated by integrating genetic association data with age- and population-specific PCa incidence and mortality statistics from the Surveillance, Epidemiology, and End Results (SEER) program and the National Center for Health Statistics (NCHS).
Key Findings
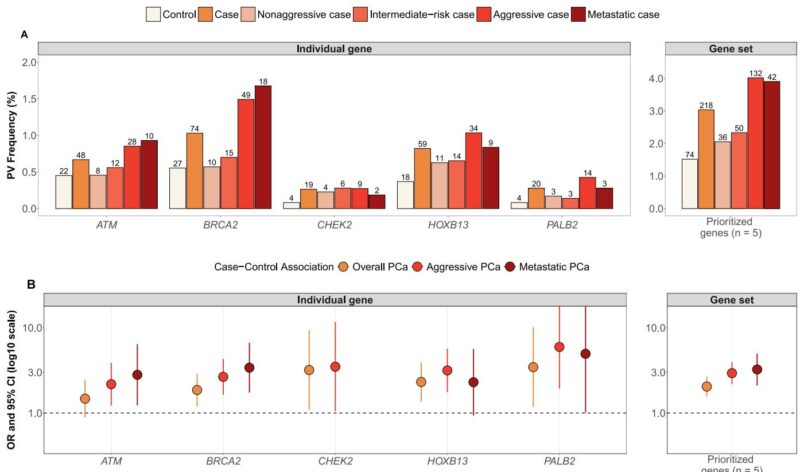
Five genes—ATM, BRCA2, CHEK2, HOXB13, and PALB2—showed significant associations with overall and particularly aggressive or metastatic PCa risk. PV frequencies in these genes were enriched among aggressive (4.0%) and metastatic (3.9%) cases compared to controls (1.5%). Odds ratios for aggressive disease ranged from 2.18 to 5.96 (p < 0.05), with carriers exhibiting markedly elevated risk relative to noncarriers.
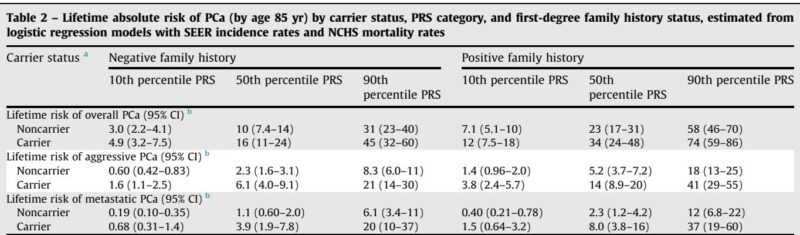
A wide spectrum of lifetime absolute PCa risk was observed depending on combined genetic and familial factors. For example, among African American men:
– Overall PCa risk ranged from 3.0% (noncarriers, low PRS, no family history) to 74% (carriers, high PRS, positive family history).
– Aggressive PCa risk spanned from 0.6% to 41%, and metastatic PCa risk varied from 0.2% to 37%.
Notably, PV carriers with a positive family history and PRS in the 90th percentile experienced sevenfold, eighteenfold, and thirty-fourfold increases in risk for overall, aggressive, and metastatic PCa, respectively, versus average-risk individuals. Age at which absolute risk thresholds were reached differed substantially; high-risk individuals reached thresholds approximately 30 years earlier than average-risk counterparts, signaling the potential benefit of earlier screening.
Additional genes—MSH5, NBN, NEIL2, and TP53—demonstrated suggestive associations, expanding the candidate gene panel for future investigation.
Expert Commentary
This comprehensive genetic analysis represents the largest of its kind in men of African ancestry, bridging critical gaps in understanding the genetic epidemiology of PCa in this population. The robust association between specific PVs and aggressive PCa aligns with prior findings in European cohorts but importantly contextualizes these within diverse genetic backgrounds.
Integrating PRS with rare variant data and family history offers superior risk stratification over traditional race-based methods, challenging current screening guidelines that recommend earlier prostate-specific antigen (PSA) testing solely on racial grounds. Precision risk estimates enable nuanced screening, potentially reducing overdiagnosis in lower-risk individuals while intensifying surveillance for those at highest risk.
The study underscores challenges inherent in rare variant research within diverse populations, such as limited power for extremely rare PVs and incomplete clinical annotation, especially from resource-limited settings. Nevertheless, consistent PV frequency patterns across multiple African countries and in African Americans enhance generalizability. Future prospective clinical trials, such as the TRANSFORM and BARCODE 1 studies, are crucial to validate the clinical utility of genetic risk-guided screening approaches.
Limitations
– Oversampling of aggressive/metastatic cases may limit direct applicability to screening populations.
– Absence of reliable incidence and mortality data from African countries restricted absolute risk modeling primarily to U.S. African American populations.
– Some misclassification of disease aggressiveness may arise when staging data were not available and PSA levels were used as proxies.
– Statistical power remained limited for some rare PVs due to their very low frequency.
Conclusions
This landmark study establishes ATM, BRCA2, CHEK2, HOXB13, and PALB2 as pivotal genes conferring increased risk of aggressive prostate cancer among men of African ancestry. The integration of rare germline pathogenic variants, polygenic risk scores, and family history refines lifetime risk estimates and highlights significant risk heterogeneity within this group.
These results advocate a paradigm shift from race-based to genetically informed, individualized PCa screening protocols. Early identification and targeted screening of high-risk men promise improved detection of aggressive disease and reduced mortality, while sparing low-risk individuals from unnecessary interventions. Comprehensive prospective trials are warranted to implement genetic risk assessment into clinical screening and prevention frameworks globally, particularly in underrepresented African populations.
AI Image Prompt
MRI prostate scan showing highlighted regions indicating aggressive prostate cancer; overlay illustrating integration of genetic data (DNA helix), polygenic risk scores (graph), and family pedigree analysis; diverse men of African ancestry silhouette in background symbolizing personalized genomic medicine.
References
Chen F, Sheng X, Wang A, Xu Y, Hughley R, Xiong W, Pooler L, Wan P, Gundell SM, Kigozi G, Nakigozi G, Nalugoda F, Kagaayi J, Kigozi GN, Mugamba S, Kyasanku E, Nkale J, Olwa VO, Lubwama A, Daama A, Nakajugo R, Adusei B, Jalloh M, Gueye SM, Adjei AA, Mensah J, Fernandez PW, Adebiyi AO, Olufemi Ogunbiyi J, Aisuodionoe-Shadrach OI, Petersen L, Chen WC, McBride J, Bensen JT, Mohler JL, Taylor JA, Andrews C, Kigongo M, Colline A, Kiddu V, Namugambe J, Owamaani S, Job K, Masaba BJ, Asiimwe F, Muwanga P, Namulondo J, Nagawa F, Kayiraba C, Ogwang M, Okidi R, Oweka D, Kitara E, Obonyo J, Lajul D, Matovu P, Muheki PA, Natumanya J, Agaba E, Aculokin E, Twongyeirwe A, Mutema G, Bitamazire D, Butler EN, Ingles SA, Rybicki BA, Stanford JL, Zheng W, Berndt SI, Chanock SJ, Huff CD, Lachance J, Multigner L, Darst BF, Rebbeck TR, Brureau L, Watya S, Conti DV, Haiman CA. Integrating Pathogenic Variants, Polygenic Risk Score, and Family History for Prostate Cancer Risk Estimation in Men of African Ancestry. Eur Urol. 2025 Nov 5:S0302-2838(25)04720-7. doi: 10.1016/j.eururo.2025.09.4161 IF: 25.2 Q1 . Epub ahead of print. PMID: 41219045 IF: 25.2 Q1 .