Highlights
– Neoadjuvant platinum chemo plus PD‑1/PD‑L1 blockade produced a 51.7% pathologic complete response (pCR) rate among 29 resectable SMARCA4‑altered NSCLCs; benefit was concentrated in squamous histology (pCR 83.3% vs 28.6% in adenocarcinoma, p=0.045).
– SMARCA4‑altered lung adenocarcinomas show heterogeneous immune phenotypes; immune‑desert cases had markedly worse survival than immune‑enriched cases.
– In advanced disease, SMARCA4 mutations enrich for higher tumor mutational burden (TMB) but overall worse median OS; co‑occurrence of STK11 and/or KEAP1 with SMARCA4 (and often KRAS) is associated with substantially shorter progression‑free and overall survival after chemoimmunotherapy.
Background
SMARCA4 encodes BRG1, a catalytic subunit of the SWI/SNF chromatin remodeling complex. Loss‑of‑function SMARCA4 alterations occur in a subset of non‑small cell lung cancer (NSCLC) and have been linked to aggressive biology, smoking association, and poor prognosis. Immune checkpoint blockade added to chemotherapy has improved outcomes broadly in resectable and advanced NSCLC, but whether SMARCA4‑altered tumors derive the same benefit—or whether outcomes differ by histology and co‑mutational context—has been unclear.
Study design and methods
This article synthesizes two recent retrospective analyses that probe the interaction of SMARCA4 alterations with neoadjuvant and advanced‑disease immunochemotherapy outcomes.
Study 1 (Peng et al., J Thorac Oncol, 2025): Single‑center (Guangdong Provincial People’s Hospital) retrospective series of 29 patients with resectable NSCLC harboring SMARCA4 alterations who received neoadjuvant immunochemotherapy. Clinical characteristics were correlated with next‑generation sequencing (NGS). The authors also interrogated The Cancer Genome Atlas (TCGA) lung adenocarcinoma cohort by SMARCA4 status (whole‑exome sequencing) and used a BostonGene molecular functional portrait to classify tumor immune microenvironment in the altered cohort.
Study 2 (Dong et al., Transl Lung Cancer Res, 2025): A cBioPortal‑based analysis of 2,098 stage IIIB–IV NSCLC cases (excluding canonical driver mutations EGFR, ALK, ROS1, RET) examining the prevalence and clinical impact of SMARCA4 mutations, and specifically the effect of STK11/KEAP1 co‑mutations on outcomes with chemoimmunotherapy. Associations with TMB, PD‑L1 expression, progression‑free survival on first‑line therapy (mPFS1), and overall survival (mOS) were evaluated with multivariable models.
Key endpoints across the studies included objective response rate (ORR), pathologic complete response (pCR) for neoadjuvant cases, event‑free survival (EFS) or progression‑free survival (PFS), and overall survival (OS). Immune classification (immune‑desert vs immune‑enriched) and pathway analyses were used to explore biological correlates.
Key findings
Neoadjuvant series (Peng et al.)
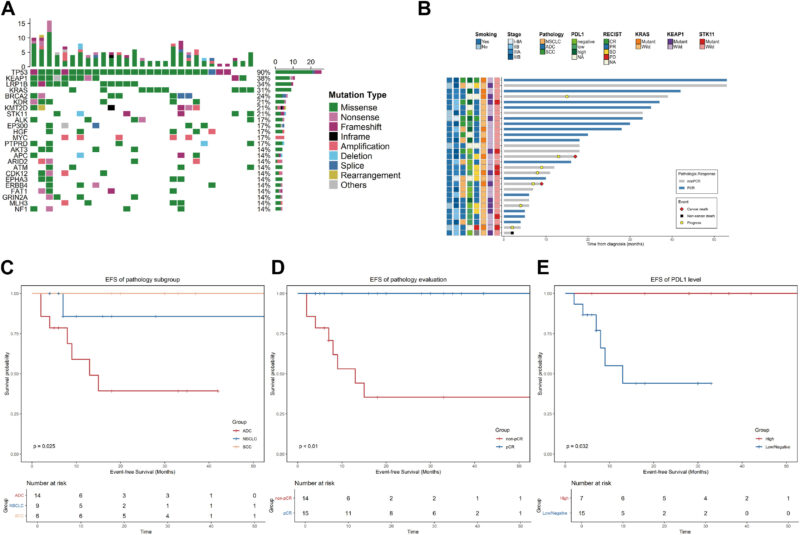
Among 29 patients with SMARCA4‑altered resectable NSCLC treated with neoadjuvant immunochemotherapy, the overall objective response rate was 70.4% and the pCR rate was 51.7%. Important subgroup findings included:
- Histology matters: pCR differed by pathology—squamous cell carcinoma (SCC) had an 83.3% pCR rate versus 28.6% for adenocarcinoma (p = 0.045).
- Relapse and survival: After a median 17‑month follow‑up, seven patients relapsed; one non‑cancer death reported. In the adenocarcinoma subgroup there was a signal for early progression (42.8%) with median EFS ≈13 months, though some adenocarcinoma patients achieved durable disease control.
- Co‑mutational risk: Patients with co‑occurring KRAS and KEAP1/STK11 alterations uniformly relapsed in this cohort, suggesting a particularly high‑risk phenotype.
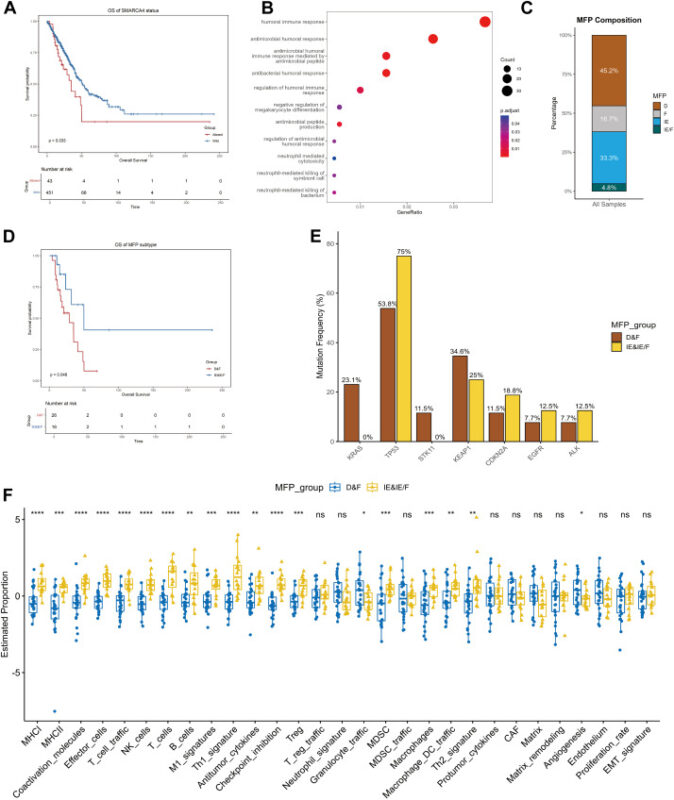
TCGA LUAD analysis mirrored the clinical observations: SMARCA4‑altered adenocarcinomas had worse survival (median 34.8 vs 50.9 months, p = 0.033) and transcriptional features consistent with down‑regulated innate immunity and enrichment for mTOR and MYC signaling pathways.
Immune microenvironment classification (BostonGene) revealed heterogeneity within SMARCA4‑altered tumors: both immune‑desert and immune‑enriched subtypes were present. Patients whose tumors lacked immune cell infiltration had significantly shorter OS than immune‑enriched cases (28.8 vs 49.9 months, p = 0.043).
Advanced disease cBioPortal analysis (Dong et al.)
Among 2,098 advanced NSCLC cases, 162 (7.7%) harbored SMARCA4 mutations. Key observations:
- Clinical phenotype: SMARCA4‑mutant patients were more commonly older, current/former smokers, and more likely to have adrenal metastases.
- Genomic features and prognosis: SMARCA4‑mutant tumors had higher TMB (P<0.001) but worse median OS (10.6 vs 17.5 months, P<0.001). SMARCA4 class I alterations carried shorter mOS and mPFS compared with class II alterations.
- Impact of adding immunotherapy: In SMARCA4‑mutant patients, first‑line chemoimmunotherapy prolonged median PFS1 compared with chemotherapy alone (5.6 vs 3.9 months, P=0.01) but did not improve mOS (10.8 vs 9.5 months, P=0.91) at the cohort level.
- Predictors of benefit: PD‑L1 positivity predicted longer mPFS1 (8.3 vs 5.1 months, P=0.02) and mOS (18.9 vs 9.3 months, P=0.03) among those receiving chemoimmunotherapy; TMB did not stratify benefit in this subgroup.
- STK11/KEAP1 co‑mutations: The presence of STK11 and/or KEAP1 alterations with SMARCA4 was not associated with TMB or PD‑L1, but significantly reduced benefit from chemoimmunotherapy—mPFS1 was 4.5 vs 13.3 months (P<0.001) and mOS 8.7 vs 20.1 months (P=0.005) compared with SMARCA4 mutants lacking these co‑mutations. Within the SMARCA4‑mutant population, patients without STK11/KEAP1 co‑mutations derived a sizable mPFS1 benefit from immunotherapy; this benefit was diminished or absent when STK11/KEAP1 co‑mutations were present.
Interpretation and mechanistic considerations
Collectively, these analyses indicate that SMARCA4‑altered NSCLC is not a single clinical entity but a spectrum defined by histology and co‑mutational context. Several mechanistic and translational points are noteworthy:
- SMARCA4 deficiency may increase genomic instability and TMB—potentially favoring immunogenicity—but this effect is overridden in subsets by co‑alterations that actively shape the immune microenvironment.
- STK11 (LKB1) loss has been associated in prior work with an immune‑cold microenvironment, impaired STING signaling, and reduced T‑cell infiltration; KEAP1 inactivation activates NRF2 and metabolic programs that can further blunt immune surveillance. The co‑presence of these lesions with SMARCA4 likely establishes an immune‑desert phenotype that is poorly responsive to immune checkpoint blockade.
- Histology‑dependent responses in the neoadjuvant setting (robust pCR in squamous cases) may reflect differences in baseline immunogenicity, tumor stromal context, or co‑mutational patterns between SCC and adenocarcinoma rather than a direct effect of SMARCA4 loss per se.
- Pathway enrichment (mTOR, MYC) identified in SMARCA4‑altered adenocarcinomas suggests alternative therapeutic vulnerabilities that could be explored in combination with immunotherapy to overcome resistance.
Expert commentary and limitations
These retrospective, hypothesis‑generating datasets provide important clinical signal but have limitations that temper immediate translation into practice:
- Size and selection bias: The neoadjuvant series included only 29 patients from a single center; outcomes may be influenced by patient selection for surgery and institutional practices.
- Retrospective design: Both studies are retrospective and observational; confounding by indication and incomplete clinical annotation (e.g., prior therapies, comorbidity, PD‑L1 assay harmonization) can affect estimates of treatment effect.
- Heterogeneity in testing and mutation classification: SMARCA4 alterations span truncating, missense, and structural events; not all alterations produce equivalent functional loss. The studies partially address this (class I vs class II), but more standardized functional annotation is needed.
- Biomarker interplay: PD‑L1 and TMB were variably informative; discordant or incomplete PD‑L1 data limit firm conclusions about its predictive role in SMARCA4‑mutant disease.
- Mechanistic inference: Transcriptomic and pathway enrichment findings are associative; prospective tissue‑based studies with single‑cell and spatial profiling are needed to confirm mechanisms of immune exclusion.
Nevertheless, the reproducible signal that STK11/KEAP1 co‑mutations identify a subgroup with poor outcomes on chemoimmunotherapy is concordant with prior literature linking these alterations—particularly in KRAS‑mutant tumors—to immune resistance. These data support the use of comprehensive NGS panels to identify SMARCA4, KRAS, STK11, and KEAP1 alterations before therapy selection.
Clinical implications and recommendations
For practicing clinicians and multidisciplinary thoracic teams, the most actionable takeaways are:
- Perform comprehensive genomic profiling (NGS) in resectable and advanced NSCLC to detect SMARCA4 and co‑occurring alterations (KRAS, STK11, KEAP1) that inform prognosis and likely responsiveness to immunotherapy.
- Neoadjuvant immunochemotherapy appears highly active in SMARCA4‑altered squamous NSCLC and can produce high pCR rates; candidate squamous patients with SMARCA4 loss may particularly benefit from this strategy pending prospective confirmation.
- In non‑squamous SMARCA4‑altered NSCLC, evaluate co‑mutational context: KRAS with STK11/KEAP1 co‑alterations mark a high‑risk, immune‑cold group for whom standard chemoimmunotherapy often yields limited benefit—these patients merit prioritized enrollment in clinical trials testing novel combinations (e.g., agents to modulate the tumor microenvironment, targeted metabolic approaches, or combinations addressing mTOR/MYC signaling).
- Consider additional immune profiling (PD‑L1, transcriptomic immune signatures, spatial immunophenotyping) where available to refine predictions for immunotherapy benefit.
Next steps for research
Prospective trials stratifying by SMARCA4 status and co‑mutational patterns are needed to validate these observations. Priority questions include:
- Can immune‑cold SMARCA4‑mutant tumors be re‑sensitized to immunotherapy using rational combinations (STING agonists, epigenetic modulators, metabolic pathway inhibitors, or NRF2/KEAP1 axis interventions)?
- Are there differences in adjuvant/neoadjuvant benefit across SMARCA4 alteration classes that could guide treatment intensity?
- What specific transcriptomic or spatial biomarkers best capture immune exclusion and predict resistance in SMARCA4‑altered NSCLC?
Conclusions
SMARCA4‑altered NSCLC is clinically and biologically heterogeneous. Current retrospective evidence indicates that squamous SMARCA4‑altered tumors are strikingly sensitive to neoadjuvant immunochemotherapy, whereas non‑squamous tumors—particularly those harboring KRAS plus STK11 and/or KEAP1 co‑mutations—comprise an immune‑cold, high‑risk subgroup with limited benefit from standard chemoimmunotherapy. These findings argue for routine comprehensive genomic profiling to guide therapeutic decision making and for the prioritization of prospective trials that test tailored combination strategies to overcome immune resistance in this challenging molecular subset.
Funding and clinicaltrials.gov
Funding and trial registration: Individual study funding and clinical trial identifiers were reported in the original manuscripts (Peng et al., J Thorac Oncol 2025; Dong et al., Transl Lung Cancer Res 2025). Readers should consult those reports for detailed acknowledgements and trial information. No additional funding statements are made in this synthesis.
References
1. Peng LS, Cui Q, Zhang C, et al. Neoadjuvant Immunochemotherapy in Resectable NSCLC With SMARCA4 Alterations. J Thorac Oncol. 2025 Oct 27. doi:10.1016/j.jtho.2025.10.013 IF: 20.8 Q1 . PMID: 41161592 IF: 20.8 Q1 .
2. Dong Z, Zuo R, Guo Y, et al. STK11/KEAP1 co-mutations in SMARCA4-mutant advanced non-small cell lung cancer: genetic characteristics and impact on immunotherapy efficacy. Transl Lung Cancer Res. 2025 Aug 31;14(8):3024-3041. doi:10.21037/tlcr-2025-305 IF: 3.5 Q1 . PMID: 40948837 IF: 3.5 Q1 ; PMCID: PMC12432681 IF: 3.5 Q1 .
3. The Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. (TCGA LUAD) — see original TCGA publications for datasets used in secondary analyses.
Note: This article synthesizes and interprets published findings. Clinicians should refer to original manuscripts for full methodological detail and institutional treatment protocols.