Introduction
Alzheimer’s disease (AD) remains a major neurodegenerative disorder characterized by progressive cognitive decline and substantial disease burden worldwide. Genetic factors critically influence AD susceptibility, among which triggering receptor expressed on myeloid cells 2 (TREM2) and apolipoprotein E (apoE) variants stand out as some of the most potent risk determinants. TREM2, a receptor on microglia, modulates immune responses and phagocytosis, while apoE, a lipid transporter with three common isoforms (apoE2, apoE3, and apoE4), affects amyloid beta metabolism and neuroinflammation. Notably, apoE4 carriers have increased AD risk and distinct pathological trajectories.
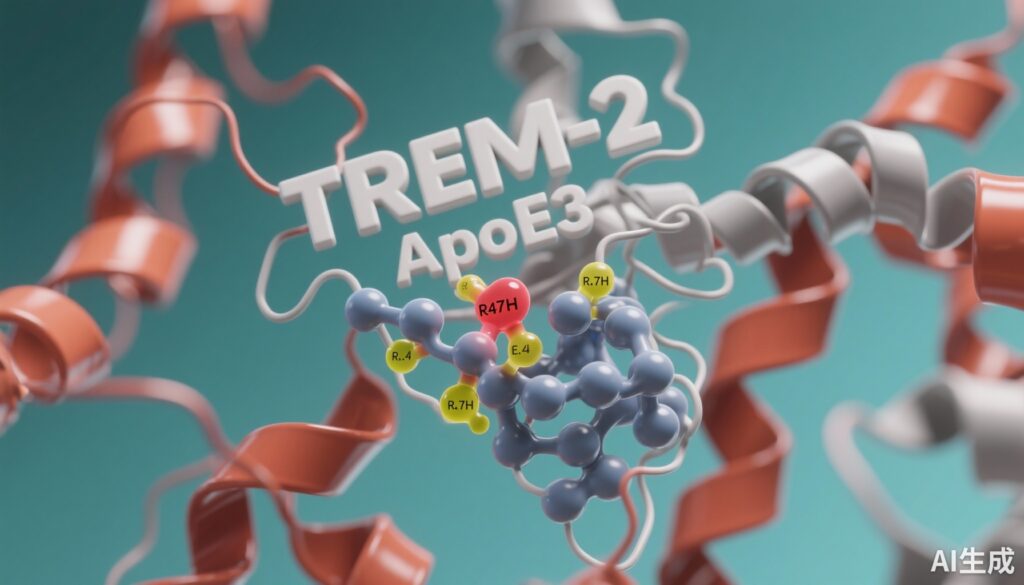
Interactions between TREM2 and apoE3 have been shown experimentally, suggesting that the molecular interplay between these proteins influences microglial functions and AD progression. However, the precise structural details of TREM2-apoE3 binding and how AD-associated variants—specifically TREM2 R47H and apoE4—alter this molecular interaction remain incompletely understood. These insights are critical for understanding pathomechanisms and for developing targeted therapies to modulate TREM2-apoE interactions in AD.
Study Design and Methods
This investigation employed computational approaches combining consensus protein-protein docking and molecular dynamics (MD) simulations. These methods aimed to generate an experimentally consistent three-dimensional complex structure of TREM2 bound to apoE3 and to interrogate the impact of AD-related variants: TREM2 R47H and apoE4. Docking identified potential interfaces and binding modes under constraints derived from experimental data, while MD simulations assessed the stability and dynamic behavior of these complexes.
The study specifically compared the conformational landscapes and interaction networks of wild-type TREM2-apoE3 complexes with those incorporating the R47H mutation on TREM2 and the apoE4 isoform in place of apoE3. Structural perturbations, binding affinities, and multimerization tendencies were analyzed to elucidate the mechanistic consequences of these mutations relevant to AD pathology.
Key Findings
The derived TREM2-apoE3 complex revealed novel interaction sites beyond previously reported contact points. These new interfaces support a more intricate molecular recognition mechanism that influences protein conformations. Binding of apoE3 induced conformational shifts in both TREM2 and apoE3, indicating a reciprocal adaptation crucial for effective interaction and potentially functional microglial signaling.
The AD-associated TREM2 R47H variant was found to alter the canonical apoE3 binding mode. Specifically, the R47H substitution shifted apoE3 binding toward a TREM2 region implicated in receptor oligomerization, potentially interfering with TREM2 multimerization and downstream signaling. This spatial shift could help explain the loss-of-function phenotype associated with R47H and its increased AD risk.
Substitution of apoE3 with apoE4 destabilized the conformational integrity of both partners in the complex. ApoE4 exhibited reduced binding stability and promoted conformational fluctuations in TREM2, which may impair receptor function and its ligand interactions. This destabilization aligns with the known increased AD risk and accelerated disease progression observed in apoE4 carriers.
Expert Commentary and Mechanistic Insights
The study’s integration of structural and dynamic analyses contributes critical mechanistic understanding of how genetic variants influence TREM2-apoE interactions. Given TREM2’s role in microglial responses to neuronal injury and amyloid pathology, disruptions in TREM2-apoE binding likely modulate microglial activation states and neuroinflammation in AD.
The observation that the R47H mutation repositions apoE3 binding toward a multimerization interface hints at a mechanism whereby this variant reduces TREM2 clustering and signaling potency. Similarly, apoE4-induced destabilization may impair ligand recognition and clearance functions. These structural perturbations provide a biologically plausible basis linking these variants to AD pathogenesis.
While compelling, these findings derive from in silico models requiring experimental corroboration through biophysical and cellular assays. Additionally, the complexity of TREM2 and apoE functions in vivo involves other cofactors and downstream pathways that warrant further investigation.
Conclusion and Clinical Implications
This study advances understanding of TREM2-apoE3 interactions at the molecular level and illustrates how AD-associated variants TREM2 R47H and apoE4 alter binding dynamics and structural stability. The identification of new interaction interfaces and conformational effects provides a foundation for rational therapeutic design aimed at restoring or modulating TREM2-apoE interactions in patients carrying these high-risk alleles.
Targeting these specific molecular defects holds promise for personalized AD interventions that could improve microglial function, reduce neuroinflammation, and slow disease progression. Future multidisciplinary research integrating structural biology, functional experiments, and clinical studies will be essential to translate these insights into effective treatments for AD.
References
Greer RA, Tuckey RA, Dean HB, Brett TJ, Roberson ED, Song Y. TREM2-apoE3 interactions and Alzheimer’s disease: Molecular and structural insights and effects of TREM2 R47H and apoE4 variants. Alzheimers Dement. 2025 Oct;21(10):e70729. doi: 10.1002/alz.70729. PMID: 41085188; PMCID: PMC12519512.